SARS-CoV-2: structures light the path to vaccines and treatment
Shape of the viral spike revealed
To gain entry into human cells, the SARS-CoV-2 virus uses a “spike” on its surface, that recognizes receptors on human cells. One approach to making a vaccine is to immunise healthy patients with DNA or mRNA that codes for the spike protein. Alternatively, it is possible to make recombinant versions of the entire spike protein, or parts of it, in the laboratory, which can then be used as vaccine candidates. Both approaches can train the immune system to produce antibodies that bind to the spike protein and block it from entering human cells. However, there are many challenges to this approach. Work on identifying vaccine candidates is underway (e.g., see here, here and here). One important question is - which parts of the protein sequence should be used in the vaccine development? This is where the structural knowledge plays a key role.
The structure of the SARS-CoV-2 spike was recently determined using electron microscopy. This revealed key molecular features such as a receptor-binding domain in a conformation available to interact with the receptor on human cells.
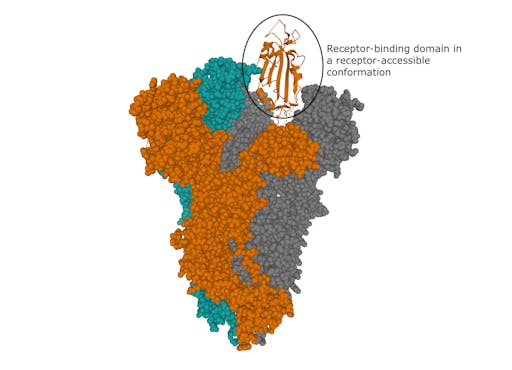
How the virus binds to human cells
How does SARS-CoV-2 gain entry into human cells? The first step in this process, the encounter between the spike protein and receptors on human cells, was revealed a few days ago by the structure determination, again by electron microscopy, of the complex between the spike and Angiotensin-converting enzyme 2 (ACE2), the cellular receptor for SARS-CoV-2. This information tells us the molecular features of the receptor that the virus uses to recognise it – knowledge that can be exploited in the design of agents that block this crucial stage of infection.
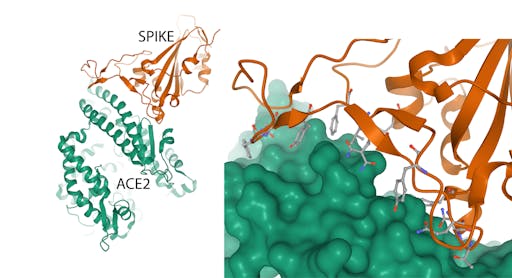
The crystal structure of an engineered receptor-binding domain in complex with human ACE2 has also been reported today.
Together, these structures represent a leap forward in the fight against COVID-19. Knowledge of the spike structure will be important for vaccine development in two key ways - selecting proteins that can be made easily in the laboratory in large amounts, and using protein engineering to enhance their stabilities, preserve their conformational properties and increase their immunogenicities (this is a nice example). Understanding how the spike binds to the ACE2 receptor on human cells paves the way to make molecules – for example antibodies – that can block this interaction by binding either the receptor-binding domain of the spike, or the receptor.
Antibodies that block viral entry
One way forward in the fight against COVID-19 is to obtain, through either immunisation strategies, or protein engineering, neutralizing antibodies that block the virus binding to human cells. The SARS-CoV-2 spike-ACE2 complex structure provides immensely powerful information to guide this approach, as it reveals the detailed molecular interactions that dictate this recognition event. So this not only informs the design of antigenic peptides for antibody, or vaccine production, but may allow rational computational design of antibodies (like here).
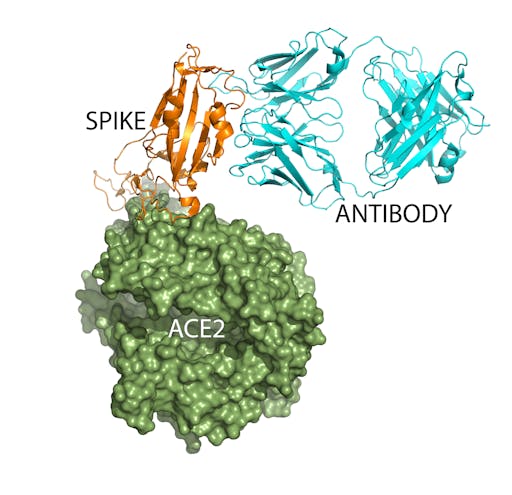
Antibodies are extremely well suited as blocking, or neutralization agents. Structural biologists have already determined the structure of a complex between the SARS-CoV-2 spike protein and a Fab portion of a human antibody. Simple structural superpositions of this complex with the spike-ACE2 complex (overlaying the spike proteins), shows that in this case while the antibody does not compete for the same binding site as ACE2 on the spike, its antigen-binding site is similar in size. This indicates that it will be possible to identify, or design neutralizing antibodies, that become powerful weapons in the fight against COVID-19. As the authors of the paper describing the crystal structure of an engineered receptor-binding domain in complex with human ACE2 point out, the receptor-binding domain could itself function as a subunit vaccine. This group have recently demonstrated this approach for the design of MERS coronavirus vaccines.
