Structural Biology of SARS-CoV-2
Like many scientific communities, structural biologists have responded extraordinarily quickly in the fight against the coronavirus (SARS-CoV-2, also referred to as 2019-nCoV) and the resultant disease COVID-19, by determining the structure of many of the viral proteins. Put simply, understanding the structures of these proteins will underpin our understanding of this virus and how to combat it with drugs and vaccines. I’ve been following the science using a few key resources that have been created especially for the crisis:
- LitCovid (Peer-reviewed COVID-19 publications)
- Protein Data Bank (PDB)
- Coronavirus Structural Taskforce
- NIH COVID-19 resource
- ViPR (Virus Pathogen Resource)
- Swiss Institute of Bioinformatics ViralZone
- EMBL-EBI Datahub
- Cell Press Coronavirus Resource Hub
- Science Magazine
- Proceedings of the National Academy of Sciences
- bioRxiv preprint server
As of writing the proteome of SARS-Co-V-2 consists of 27 proteins (see annotation).
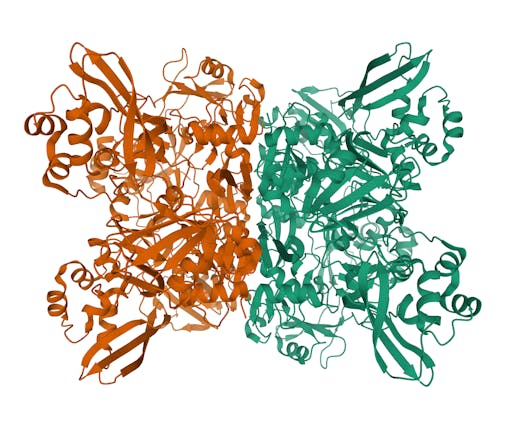
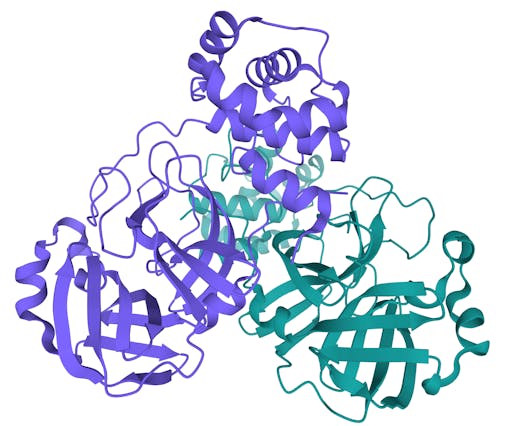
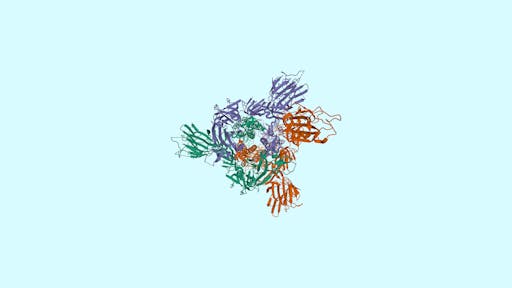
Over the past week the number of COVID19-related structures deposited in the Protein Data Bank (PDB) has grown extremely quickly - currently there are 104 structures deposited. Impressively, there are 69 structures for the main protease in complex with ligands or inhibitors. This is important because it represents a crucial step in the process of rational drug design – more on that in a later post.
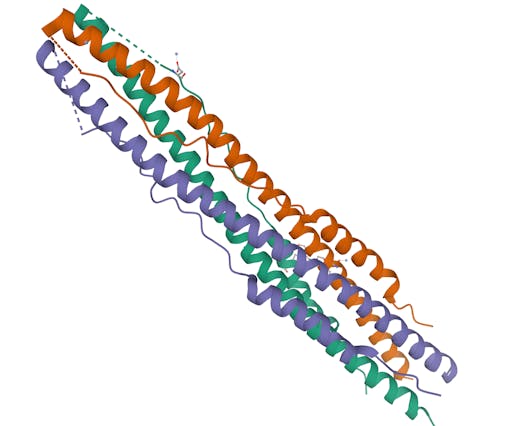
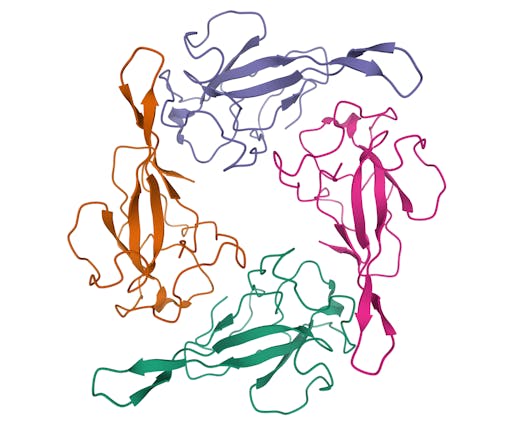
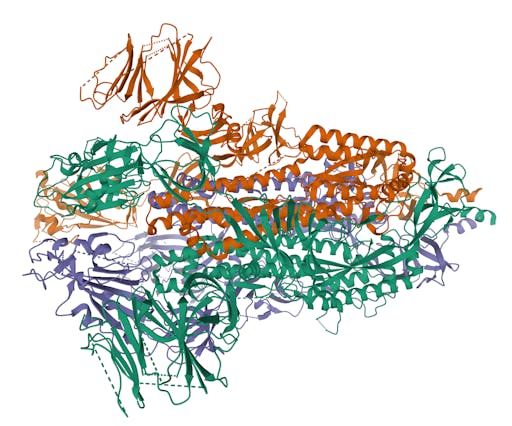
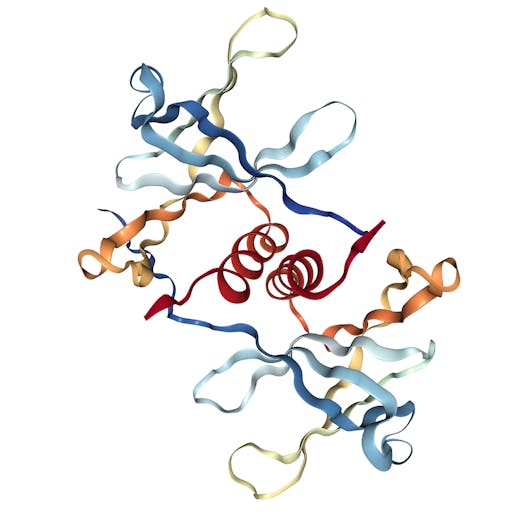
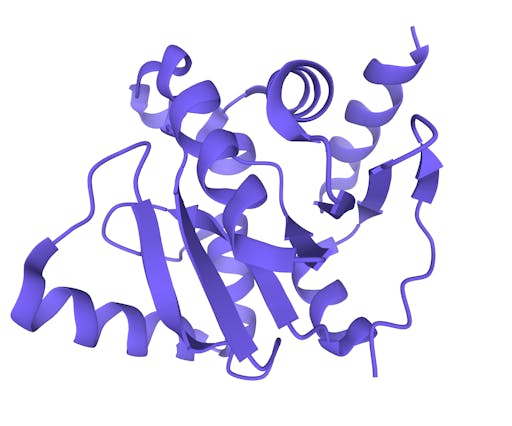
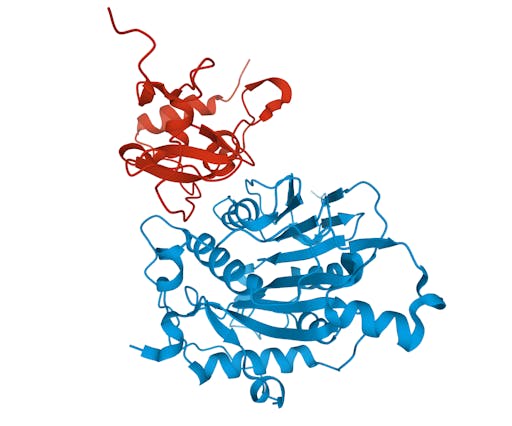
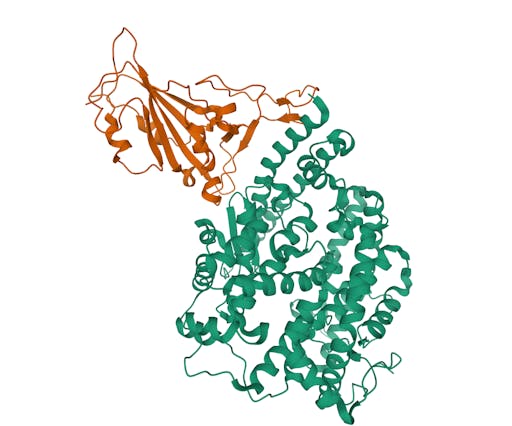
As one might expect, the vast majority (100) of these are from X-ray crystallographic data, however 4 are from Electron microscopy (EM) – this is testament to the “resolution revolution” and the emergence of EM as a powerful tool in structural biology. All of these EM structures are of the virus spike glycoprotein, which comprises the receptor binding domain and is thus the molecule that targets the receptors on human cells. This in an important piece of the puzzle – structural knowledge of this molecule is key to making vaccines, and also antibodies that might neutralize the virus. More on this in an upcoming post. For now, here is a gallery showing the different folds represented by COVID19-related structures deposited in the PDB so far: